Testing project pages
This paper is a review of an emerging trend in scientific data distribution, but one that is not emerging fast enough. It is a bit different from the other papers described on this site as it is a review rather than a piece of original research, but the topic it discusses - web APIs and how they should be used to distribute scientific data - underpins a lot of modern research, especially in Bioinformatics. As always, click the link above to see the full paper.
Data Access in Science
When we think of scientific research, we might imagine teams of people working in labs, wearing lab coats, delicately transferring mysterious (and eye wateringly expensive) quantities of liquid from vial to another. Or we might think of incredibly complex (and again, staggeringly expensive) machines making strange noises as they perform some task human hands are far too clumsy to carry out.
And for the most part, that is where a lot of the science that underpins modern advances is done. But less obvious is the more intangible stuff - the data produced by all that work. Whenever an experiment is run, data is produced in the form of measurements, or analysis on those measurements, or analysis on that analysis, and so on. It's this data which lets you make inferences about the world. Without the data and its analysis, all the work done in the lab is for nothing.
But modern science is collaborative and global. If every research team produced their own data and just analysed that, we would needlessly duplicate effort on a colossal scale. We would also be limited by the ideas and expertise of the lab that produced it, rather than letting the whole world (in practice, other people in that field) examine it. Science relies on data produced by one lab being made available to anyone. And not in a 'let me know if you're interested in what we're doing and I'll email you some files when I get around to it' - systematised, reliable, automated availability.
This is a particular problem in Biology. A lot of research depends on things like sequencing entire genomes, or calculating the three dimensional structures of proteins, and that generates a lot of data - think hundreds of gigabytes at a minimum, and often much more. This is not data that you can email to your colleague in another lab as an email attachment, it is enormous. Dealing with the problem of how this data, which is central to so many other experiments, can be accessed by others, was a big headache in the 1980s and 1990s.
Part of the solution lies in having centralised organisations which act as a single data source. For example in Structural Biology there is an organisation called the Protein Data Bank. When a lab works out the 3D coordinates of a new protein, they don't invite the whole world to download that structure from them and their servers, they send it to the Protein Data Bank, and they make it available on their dedicated infrastructure.
But size is not the only constraint on making this data available. When the web started to become widely adopted in the mid-1990s, many organisations set up webpages and webforms written in HTML, where humans could navigate to a page, and manually download some data file - perhaps with a form for providing options or running a 'job' of some kind. Now anyone who wanted some data can manually go to a web page, navigate whatever links and forms are in place, and download the data files to their computer.
And this is fine if you just want that data, once. But it's a manual process. A human has to manually navigate a web page every time data is to be downloaded. Often though, you need to be able to automate this process - to download the data every few days for example, or by writing a computer program which downloads the data as part of its operation to feed into some other pipeline. Computer programs struggle to read and navigate web pages reliably, and if data was only available via web pages, this would be like doing Science with the handbrake on.
Web APIs
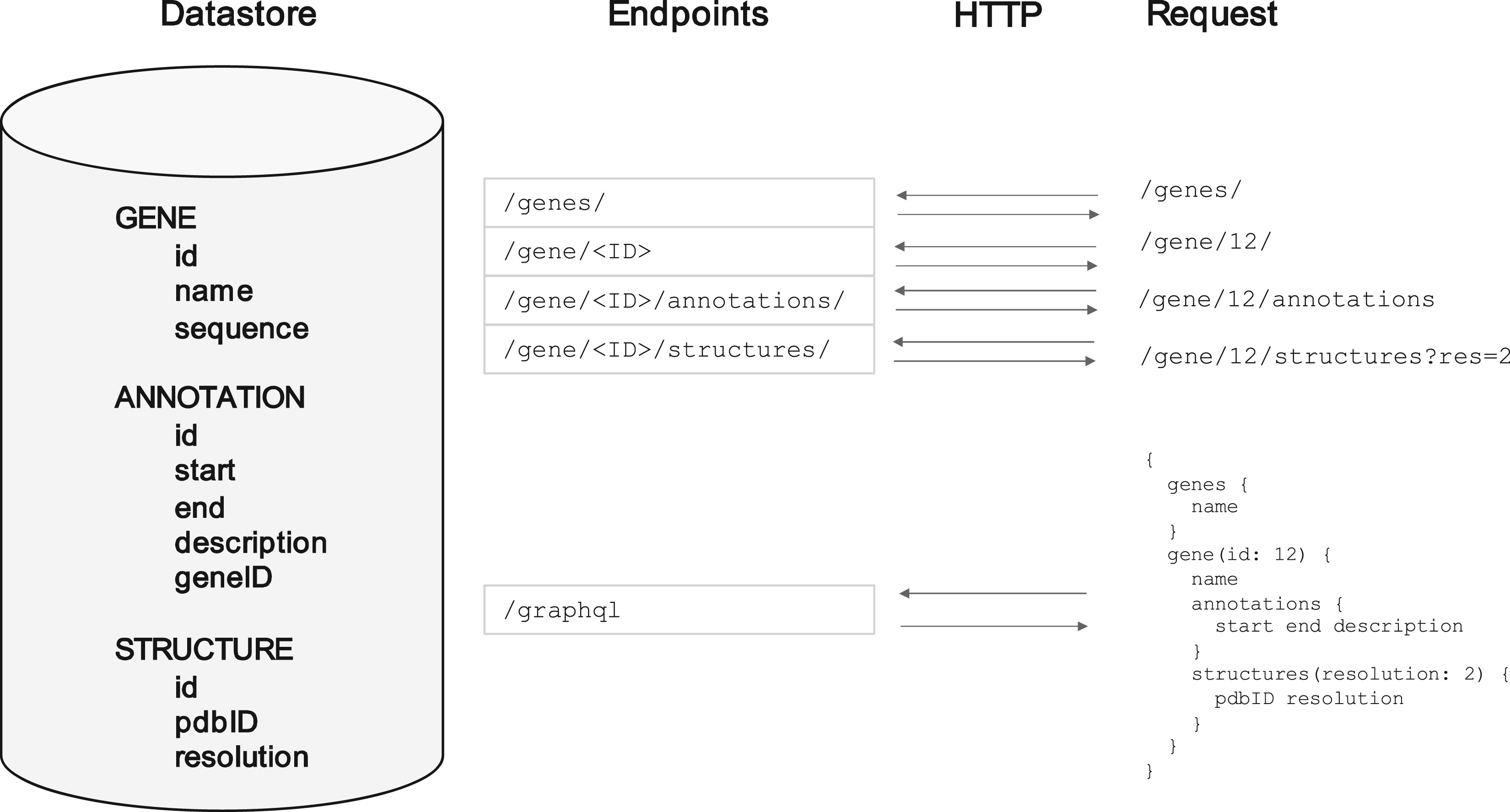
This is the problem that 'web APIs' solve. They work in a similar way to the normal web that humans interact with, but instead of a human typing an address into a browser and getting a HTML page back, a bot or program will request some resource at some address, and get data back in a strict format like JSON or XML, which they know how to interpret. HTML is the language that describes how web pages should look and there's a near infinite ways of laying out a page and interface. JSON and XML however are strict data descriptive languages - they can be read in an automated way by a computer program.
A common pattern that was arrived at early on was the 'REST API'. This works very similarly to how browsers work. Rather than each page having its own unique address, each 'resource' has its own unique address. So to get the data on one gene, you might send a request to something at http://api.big-organisation.org/genes/g101 where 'g101' is that gene's unique ID. Instead of getting a HTML web page back with buttons and pictures on it, you a representation of the gene in a computer-readable form such as JSON (see diagram below). A computer program can automatically send a request for this, parse the contents easily, and do whatever analysis on it that it needs to. If all it had was the HTML, it would not be able to extract the data it needed unless a human explicitly looked at the HTML first and marked down where the key information was (which could change at any time).
Testing captions